Hybrid OpenMP and MPI within the CASTEP code
eCSE01-017Key Personnel
PI/Co-I: Dr Matt Probert, Dr Phil Hasnip - University of York; Prof Keith Refson - Royal Holloway, University of London; Dr Ian Bush - Oxford e-Research Centre
Technical: Edward Higgins - University of York
Relevant Documents
eCSE Technical Report: Hybrid OpenMP and MPI within the CASTEP code
Project summary
Methods for performing first-principles simulations of materials, i.e. those that solve the quantum-mechanical Schrödinger equation via parameter-free approximations, have had a profound and pervasive impact on science and technology, spreading from physics, chemistry and materials science to diverse areas including electronics, geology and medicine. Methods based on density functional theory (DFT) have led the way in this success, offering a favourable balance of computational cost and accuracy. However, DFT is certainly not a computationally "cheap" method and it is essential to optimise codes not only for single-core performance but also for large parallel calculations.
CASTEP is a DFT code developed in the UK and is commonly used to model systems of up to a few thousand atoms on ARCHER using up to a few thousand cores (with larger systems scaling well to larger number of cores). It has consistently been in the top 10 codes by core hours across both HECToR and ARCHER, using over 5% of the machine over the course of a year. CASTEP was rewritten in 1999-2001 and has been actively developed since then - and this project lays the foundations for a version of CASTEP that will work well on future exascale supercomputers.
For very large systems (e.g. more than 5000 electrons) the amount of replicated data on each CPU core previously exceeded the memory available to that core. This project added a new level of parallelism to CASTEP, allowing memory to be shared between CPU cores. This allows systems of over 4x more electrons to be simulated on ARCHER than was possible before the project, allowing larger and more complex systems to be studied than before. An illustration of the memory saving for a demonstration calculation on the crambin protein residue (3660 electrons) is given in the figures below:

Crambin unit cell
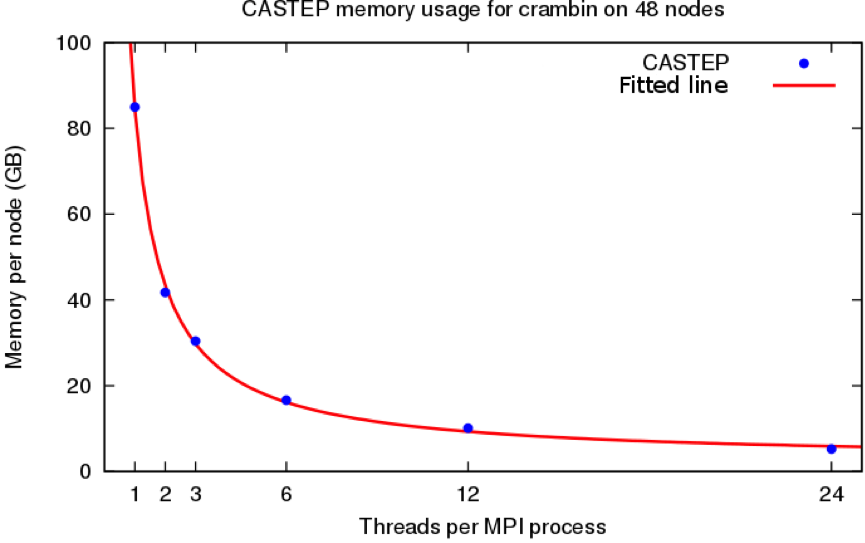
Memory saving for crambin on 48 nodes. Pre-CASTEP v16.1,
only 1 thread per MPI process was possible
In addition to this, some tasks in CASTEP were performed in serial on all cores. While most of this work takes a small portion of the runtime, larger systems are starting to take significant time in some of these tasks. This project aimed to distribute the work in these significant tasks across multiple cores, therefore significantly reducing the time taken.
CASTEP is currently used by over 850 academic research groups worldwide and so it has a large user-base. Within the UK, it is used a lot by the UKCP and the Materials Chemistry HEC consortia and by members of CCP-NC for crystalline NMR simulations. This user base spans a wide range of materials research in Physics, Chemistry, Materials Science, Earth Sciences and Engineering departments. The success of this project means that CASTEP can now be used for larger system sizes on ARCHER than ever before; that calculations that were near the memory limit before can now run significantly faster on the same number of nodes; and that large calculations can now scale to larger core counts for an even better speed up. The net result is a new science capability, running larger system sizes in less time.
Summary of the software
CASTEP (http://www.castep.org) is a UK-based state-of-the-art implementation of DFT and a flagship code for UK HPC. It was rewritten in 1999-2001 according to sound software engineering principles and with HPC in mind. CASTEP describes the electronic states ("bands") of a material using a plane-wave (Fourier) basis, necessitating the heavy use of parallel fast Fourier transforms (FFTs) for efficiency. This implementation, while general, is very well-suited to solid-state applications including simulations of materials containing defects and interfaces.
CASTEP is currently available as a system binary on ARCHER, and will be updated to v16.1, which includes this work, shortly after its release later this year. In addition to this, CASTEP is available under a free of charge licence to UK academic research groups and source code licence can be purchased by European academics for €1800. For industry and the rest of the world, CASTEP can be purchased through BIOVIA. For more information, see http://castep.org/CASTEP/GettingCASTEP.






